NB: this series is still a work in progress.
This post builds off of our previous discussions on healthcare AI infrastructure. It may be helpful to review the posts that cover the general lay of healthcare IT land, development infrastructure, and implementation infrastructure.
C. difficile Infection Model
We will be discussing the technical integration of a model that we have running at the University of Michigan. We developed this model with the intent to C. difficile infection risk stratification
This is the model that we developed to integrated for C. difficile infection risk stratification.
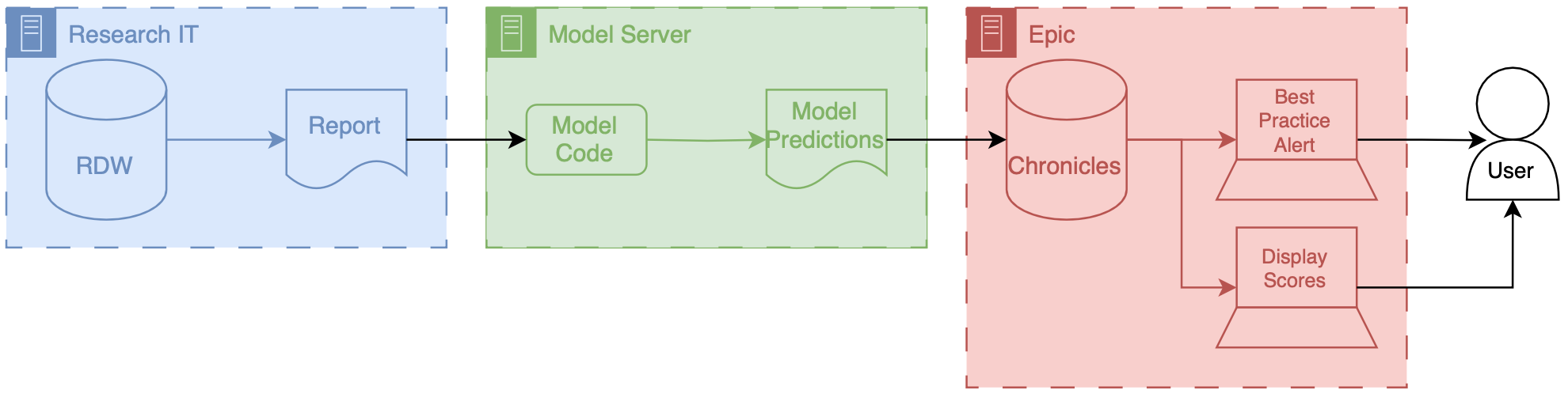
Data for this model comes from our research data warehouse then travels to the model posted on a Windows virtual machine. The predictions from the model are then passed back to the EMR using web services.
We have a report that runs daily from the research state warehouse. It’s a stored SQL procedure that runs at a set time very early in the morning about 5 AM. This is essentially a large table of data for each of the patients that were interested in producing a prediction on rows our patients and columns are the various features that were interested in. Stored procedures update information in a view inside of RDW.
The research data at warehouse and this view are accessible by a Windows machine that we have inside of the health IT secure computing environment. This windows machine has a scheduled job that runs every morning about at about 6 AM. This job pull the data down from the database runs a series of python files that you data pre-processing and apply the model to the data to the transform data, and then save the output, model predictions to a shared secured directory on the internal health system network.
We then returned the predictions to Chronicles, using infrastructure that our health IT colleagues helped to develop. This infrastructure involves a scheduled job written in C# that reads the file that we have saved the shared directory does date of validation and then passes data into chronicles using epics web services framework.
These data end up as flow sheet values for each patient. We then worked with our epic analyst colleagues to use the flow sheet data to trigger as practice alerts, and also to populate port. The best practice alerts fire based off of some configuration that’s done inside of epic in order to be able to adjust the alerting threshold outside of Epic what we did was we modified the score such that the alerting information with someone distinct from the actual score so what we did is we packed an alert flag and the score together into a single decimal separated value and this is essentially a number however it’s unique and that it contains two pieces of information so we could take a patient to Oehlert on and we would say 1.56 a patient that we didn’t alert on would be zero point
Model predictions are passed to the EMR system using web services. Predictions are then filed as either flowsheet rows (inpatient encounters) or smart data elements (outpatient encounters). You have to build your own infrastructure to push the predictions to the EMR environment.
Cheers,
Erkin
Go ÖN Home

